Patient-specific Analysis of Left Ventricle Motion
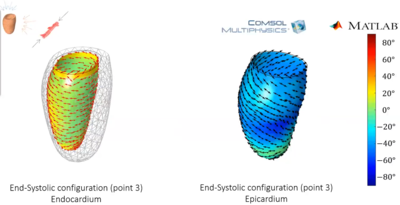
We present a framework for patient-specific study of cardiac motion; in particular, we focus on the deformations of a beating Left Ventricle (LV), confronting numerical simulations with real data acquired by echo-cardiography. Our goal is to determine the clinical importance of the LV strains pattern and to investigate its relationship with the arrangement of myocardial fibers. The proposed framework could in principle be used for a wide range of clinical applications. The fibrous composition of the myocardium is well recognized; nevertheless, a detailed description of the fibers arrangement is unknown. Its architecture suggests that muscular fibers are a major constituents of myocardial wall and that strains should be oriented in the same direction as the fibers; for these reasons, the circumferential and longitudinal strains are very much used to assess the myocardial function. Recent studies have suggested that the strains may help revealing real fiber orientation and could eventually be used to detect some pathologies (1,2).. In vivo analysis of heart is a challenging task. On the one hand, in vivo imaging techniques cannot provide accurate information on the actual architecture of the heart. On the other hand, anatomical studies provide detailed tissue descriptions of a steady heart. We tackle this problem by confronting the real motion as detected by 3D Speckle Tracking Echocardiography (3DSTE) with the motion of a simulated LV, following the method proposed in (3) to compute the strains. Our algorithm is as follows: for any subject, 3DSTE can record about 20 shapes of the beating LV during the cardiac revolution. The diastolic shape is used as patient-specific reference shape, is used in COMSOL Multiphysics® as computational domain and is endowed with the physics apt to describe myocardial contractions. Then, the shapes produced by the simulations are confronted with the actual ones, and the parameters of the COMSOL Multiphysics® model are tuned so that, eventually, the real motion is reproduced. To achieve our goal, we introduce a computational representation of the LV that captures the passive and active behavior of the cardiac tissue as well as the fiber architecture (see panel A in Figure 1) using the Equation-Based Modeling interface of COMSOL Multiphysics®. Moreover, the simulation of the ventricular function results from the solution of a series of mechanical problems that demand a fine-tuning of modeling parameters with a special emphasis on the active response. This is not a trivial task; here, we take advantage of the LiveLink™ for MATLAB® interface to extend our modeling with scripting programming in the MATLAB® environment.
References 1. Gabriele S., Nardinocchi P., Varano V. Evaluation of the strain-line patterns in a human ventricle: a simulation study. Computer methods in biomechanics and biomedical engineering. 18(7): 790-798 (2015). 2. Lee, S., Choi, S., Kim, S., Jeong, Y., Lee, K., Hur, S. H.,…& Song, J. M. Validation of three‐dimensional echocardiographic principal strain analysis for assessing left ventricular contractility: An animal study. Medical physics, 46(5), 2137-2144 (2019). 3. Pedrizzetti G., et al. Functional strain patterns in the human left ventricle. Physical review letters. 109(4):048103 (2012)